11. Design of cryptographically robust Vector Boolean functions¶
11.2. Optimization of Boolean functions via Genetic Algorithms¶
11.2.1. Description¶
11.2.1.1. Galib¶
The Genetic Algorithm Library called GAlib was (straightforwardly) linked with our VBF library to perform a search of Boolean functions with good combined cryptographic criteria. In using the GAlib library we will work primarily with two classes: a genome and a genetic algorithm. Each genome instance represents a single solution to our optimization problem. The genetic algorithm object defines how the evolution should take place. The genetic algorithm uses an objective function to determine how ‘fit’ each genome is for survival. It uses the genome operators (built into the genome) and selection/replacement strategies (built into the genetic algorithm) to generate new individuals.
The following three items must be defined in order to solve an optimization problem using a genetic algorithm: A representation, the genetic operators and the objective function.
The genetic algorithm object determines which individuals should survive, which should reproduce, and which should die. It also records statistics and decides how long the evolution should continue. The algorithm updates the population of solutions over a number of iterations (or generations*). We have used the number of generations as a stopping measure. In each iteration a number of steps are involved:
- Selection of parents from the current population of solutions.
- Crossover of parents to produce offspring.
- Mutation of the offspring.
- Selection from the mutated offspring and the current population of solutions to determine the population of solutions for the next iteration.
Among the many different types of genetic algorithms offered by GAlib we have chosen the standard ‘simple genetic algorithm’ described by Goldberg in his book [Goldberg:1989]. This algorithm uses non-overlapping populations and optional elitism. Each generation the algorithm creates an entirely new population of individuals.
11.2.1.2. Representation¶
When you use a genetic algorithm to solve an optimization problem, you must be able to represent a single solution to your problem in a single data structure. The genetic algorithm will create a population of solutions based on a sample data structure that you provide. The genetic algorithm then operates on the population to evolve the best solution. In GAlib, the sample data structure is called a GAGenome (some people refer to it as a chromosome). We have used a type of genome called GA2DBinaryStringGenome. This class is derived from the base GAGenome class and a data structure class which consists of a 2-dimensional array of Boolean with 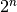 elements (the binary string is the Truth Table of the Boolean function).
elements (the binary string is the Truth Table of the Boolean function).
11.2.1.3. Genetic operators¶
Each genome has three primary operators: initialization, mutation, and crossover. With these operators you can bias an initial population, define a mutation or crossover specific to our representation, or evolve parts of the genetic algorithm as our population evolves.
The initialization operator determines how the genome is initialized. It is called when you initialize a population or the genetic algorithm. This operator does not actually create new genomes, rather it ‘stuffs’ the genomes with the primordial genetic material from which all solutions will evolve. We have used a uniform random initialization operator.
The mutation operator defines the procedure for mutating each genome. The mutation operation introduces randomness to the population of solutions. Mutation is generally applied to the children which result from the breeding process. We have used the typical mutator for a binary string genome which flips the bits in the string with a given probability (uniform random bit flip).
The crossover operator defines the procedure for generating a child from two parent genomes in order to obtain offspring. The crossover operation involves selecting two “parents” from the current population of solutions, picking a random point in the binary string representing each of the parents and swapping the values beyond that point between the two parents. This process results in two “children” with some characteristics of each of the parents.
11.2.1.4. Weighted Objective Functions¶
In addition to the three primary operators, each genome must also contain an Objective Function. The Objective Function is used to evaluate the genome in order to know how good it is compared to the other genomes. Several objective functions were employed gradually involving more criteria in a weighted manner:
1. Nonlinearity of the Boolean function: 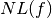 . This cryptographic criterion is represented by a locally smooth fitness
function:
. This cryptographic criterion is represented by a locally smooth fitness
function:
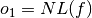
- The sum of the nonlinearity and linearity distance of the Boolean function, normalized with respect to their (a priori known) maximum values:
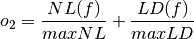
where maxNL and maxLD are the maximum values of nonlinearity and linearity distance which can be achieved by a Boolean function with the same number of input variables as f respectively.
- The sum of nonlinearity, algebraic degree and linearity distance of the Boolean function normalized with respect to their (a priori known) maximum values:
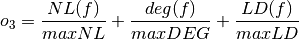
where maxNL, maxDEG and maxLD are respectively the maximum values of nonlinearity, algebraic degree and linearity distance which can be achieved by a Boolean function with the same number of input variables as f.
- The sum of nonlinearity, algebraic degree, algebraic immunity and linearity distance of the Boolean function normalized with respect to their (a priori known) maximum values:
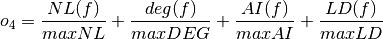
where maxNL, maxDEG, maxAI and maxLD are respectively the maximum values of nonlinearity, algebraic degree, algebraic immunity and linearity distance which can be achieved by a Boolean function with the same number of input variables as f.
- The sum of nonlinearity, algebraic degree, algebraic immunity and linearity distance of the Boolean function normalized with respect to their (a priori known) maximum values:
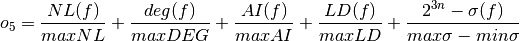
where maxNL, maxDEG, maxAI, maxLD, 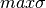 are respectively the maximum values of nonlinearity, algebraic degree, algebraic immunity, linearity distance and sum-of-square indicator which can be achieved by a Boolean function with the same number of input variables as $f$; and
are respectively the maximum values of nonlinearity, algebraic degree, algebraic immunity, linearity distance and sum-of-square indicator which can be achieved by a Boolean function with the same number of input variables as $f$; and 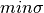 is the minimum value of the sum-of-square indicator achievable by a Boolean function with the same number of input variables as f.
is the minimum value of the sum-of-square indicator achievable by a Boolean function with the same number of input variables as f.
11.2.2. Implementation¶
In order to use Galib library, the installation instructions described in Installation Instructions for GAlib must be followed. Once Galib is installed, we can use this library in conjunction with VBF library by using the following Makefile:
include makevars
############# User settings ######################
# set your C++ compiler and options here...
GPP=g++
LIBS=-lntl
NTLINC= -I/usr/local/include -L/usr/local/lib
GA_LIB_DIR= ga
LIB_DIRS= -L$(GA_LIB_DIR)
############# End of user settings ###############
o1: o1.cpp VBF.h
$(GPP) $(NTLINC) -Wall o1.cpp -o o1.exe $(LIBS) $(LIB_DIRS) -lga
o2: o2.cpp VBF.h
$(GPP) $(NTLINC) -Wall o2.cpp -o o2.exe $(LIBS) $(LIB_DIRS) -lga
o3: o3.cpp VBF.h
$(GPP) $(NTLINC) -Wall o3.cpp -o o3.exe $(LIBS) $(LIB_DIRS) -lga
o4: o4.cpp VBF.h
$(GPP) $(NTLINC) -Wall o4.cpp -o o4.exe $(LIBS) $(LIB_DIRS) -lga
o5: o5.cpp VBF.h
$(GPP) $(NTLINC) -Wall o5.cpp -o o5.exe $(LIBS) $(LIB_DIRS) -lga
clean:
rm -f *.exe
Note that a file called “makevars” must be present in the same directory as the Makefile file.
Example program for each of the objective functions could be the following:
#include <ga/GASimpleGA.h> // we're going to use the simple GA
#include <ga/GA2DBinStrGenome.h> // and the 2D binary string genome
#include <ga/std_stream.h>
#include "VBF.h"
#define cout STD_COUT
float Objective(GAGenome &); // This is the declaration of our obj function.
int
main(int argc, char **argv)
{
int n = 0, m = 0;
int popsize;
int ngen;
float pmut;
float pcross;
// We generate random seed
for(int ii=1; ii<argc; ii++) {
if(strcmp(argv[ii++],"seed") == 0) {
GARandomSeed((unsigned int)atoi(argv[ii]));
}
}
n = atoi(argv[1]);
m = atoi(argv[2]);
int width = 1 << n;
int height = m;
popsize = atoi(argv[3]);
ngen = atoi(argv[4]);
pmut = atof(argv[5]);
pcross = atof(argv[6]);
GA2DBinaryStringGenome genome(width, height, Objective);
GASimpleGA ga(genome);
ga.populationSize(popsize);
ga.nGenerations(ngen);
ga.pMutation(pmut);
ga.pCrossover(pcross);
ga.evolve();
// Now we print out the best genome that the GA found.
cout << ga.statistics() << "\n";
cout << ga.statistics().bestIndividual() << "\n";
return 0;
}
float
Objective(GAGenome& g) {
using namespace VBFNS;
VBF F;
NTL::mat_GF2 T;
NTL::RR nonlin;
long spacen, m;
GA2DBinaryStringGenome & genome = (GA2DBinaryStringGenome &)g;
float score=0.0;
spacen = genome.width();
m = genome.height();
T.SetDims(spacen,m);
for(int i=0; i<spacen; i++){
for(int j=0; j<m; j++){
T[i][j] = to_GF2(genome.gene(i,j));
}
}
F.puttt(T);
nonlin = nl(F);
conv(score,nonlin);
return score;
}
#include <ga/GASimpleGA.h> // we're going to use the simple GA
#include <ga/GA2DBinStrGenome.h> // and the 2D binary string genome
#include <ga/std_stream.h>
#include "VBF.h"
#define cout STD_COUT
float Objective(GAGenome &); // This is the declaration of our obj function.
int
main(int argc, char **argv)
{
int n = 0, m = 0;
int popsize;
int ngen;
float pmut;
float pcross;
// We generate random seed
for(int ii=1; ii<argc; ii++) {
if(strcmp(argv[ii++],"seed") == 0) {
GARandomSeed((unsigned int)atoi(argv[ii]));
}
}
n = atoi(argv[1]);
m = atoi(argv[2]);
int width = 1 << n;
int height = m;
popsize = atoi(argv[3]);
ngen = atoi(argv[4]);
pmut = atof(argv[5]);
pcross = atof(argv[6]);
GA2DBinaryStringGenome genome(width, height, Objective);
GASimpleGA ga(genome);
ga.populationSize(popsize);
ga.nGenerations(ngen);
ga.pMutation(pmut);
ga.pCrossover(pcross);
ga.evolve();
// Now we print out the best genome that the GA found.
cout << ga.statistics() << "\n";
cout << ga.statistics().bestIndividual() << "\n";
return 0;
}
float
Objective(GAGenome& g) {
using namespace VBFNS;
VBF F;
NTL::mat_GF2 T;
NTL::RR nonlin, lind, suma, nlm, ldm;
long spacen, m;
GA2DBinaryStringGenome & genome = (GA2DBinaryStringGenome &)g;
float score=0.0;
spacen = genome.width();
m = genome.height();
T.SetDims(spacen,m);
for(int i=0; i<spacen; i++){
for(int j=0; j<m; j++){
T[i][j] = to_GF2(genome.gene(i,j));
}
}
F.puttt(T);
nonlin = nl(F);
lind = ld(F);
nlm = nlmax(F);
ldm = ldmax(F);
suma = nonlin/nlm+lind/ldm;
conv(score,suma);
return score;
}
#include <ga/GASimpleGA.h> // we're going to use the simple GA
#include <ga/GA2DBinStrGenome.h> // and the 2D binary string genome
#include <ga/std_stream.h>
#include "VBF.h"
#define cout STD_COUT
float Objective(GAGenome &); // This is the declaration of our obj function.
int
main(int argc, char **argv)
{
int n = 0, m = 0;
int popsize;
int ngen;
float pmut;
float pcross;
// We generate random seed
for(int ii=1; ii<argc; ii++) {
if(strcmp(argv[ii++],"seed") == 0) {
GARandomSeed((unsigned int)atoi(argv[ii]));
}
}
n = atoi(argv[1]);
m = atoi(argv[2]);
int width = 1 << n;
int height = m;
popsize = atoi(argv[3]);
ngen = atoi(argv[4]);
pmut = atof(argv[5]);
pcross = atof(argv[6]);
GA2DBinaryStringGenome genome(width, height, Objective);
GASimpleGA ga(genome);
ga.populationSize(popsize);
ga.nGenerations(ngen);
ga.pMutation(pmut);
ga.pCrossover(pcross);
ga.evolve();
// Now we print out the best genome that the GA found.
cout << ga.statistics() << "\n";
cout << ga.statistics().bestIndividual() << "\n";
return 0;
}
float
Objective(GAGenome& g) {
using namespace VBFNS;
VBF F;
NTL::mat_GF2 T;
NTL::RR nonlin, lind, suma, nlm, ldm;
long spacen, m;
int d, n;
GA2DBinaryStringGenome & genome = (GA2DBinaryStringGenome &)g;
float score=0.0;
spacen = genome.width();
m = genome.height();
T.SetDims(spacen,m);
for(int i=0; i<spacen; i++){
for(int j=0; j<m; j++){
T[i][j] = to_GF2(genome.gene(i,j));
}
}
F.puttt(T);
nonlin = nl(F);
lind = ld(F);
nlm = nlmax(F);
ldm = ldmax(F);
d = deg(F);
n = logtwo(spacen);
suma = nonlin/nlm+lind/ldm+to_RR(d)/to_RR(n);
conv(score,suma);
return score;
}
#include <ga/GASimpleGA.h> // we're going to use the simple GA
#include <ga/GA2DBinStrGenome.h> // and the 2D binary string genome
#include <ga/std_stream.h>
#include "VBF.h"
#define cout STD_COUT
float Objective(GAGenome &); // This is the declaration of our obj function.
int
main(int argc, char **argv)
{
int n = 0, m = 0;
int popsize;
int ngen;
float pmut;
float pcross;
// We generate random seed
for(int ii=1; ii<argc; ii++) {
if(strcmp(argv[ii++],"seed") == 0) {
GARandomSeed((unsigned int)atoi(argv[ii]));
}
}
n = atoi(argv[1]);
m = atoi(argv[2]);
int width = 1 << n;
int height = m;
popsize = atoi(argv[3]);
ngen = atoi(argv[4]);
pmut = atof(argv[5]);
pcross = atof(argv[6]);
GA2DBinaryStringGenome genome(width, height, Objective);
GASimpleGA ga(genome);
ga.populationSize(popsize);
ga.nGenerations(ngen);
ga.pMutation(pmut);
ga.pCrossover(pcross);
ga.evolve();
// Now we print out the best genome that the GA found.
cout << ga.statistics() << "\n";
cout << ga.statistics().bestIndividual() << "\n";
return 0;
}
float
Objective(GAGenome& g) {
using namespace VBFNS;
VBF F;
NTL::mat_GF2 T;
NTL::RR nonlin, lind, suma, nlm, ldm;
long spacen, m;
int d, n, ai, maxai;
GA2DBinaryStringGenome & genome = (GA2DBinaryStringGenome &)g;
float score=0.0;
spacen = genome.width();
m = genome.height();
T.SetDims(spacen,m);
for(int i=0; i<spacen; i++){
for(int j=0; j<m; j++){
T[i][j] = to_GF2(genome.gene(i,j));
}
}
F.puttt(T);
nonlin = nl(F);
lind = ld(F);
nlm = nlmax(F);
ldm = ldmax(F);
d = deg(F);
n = logtwo(spacen);
ai = AI(F);
maxai = aimax(F);
suma = nonlin/nlm+lind/ldm+to_RR(d)/to_RR(n)+to_RR(ai)/to_RR(maxai);
conv(score,suma);
return score;
}
#include <ga/GASimpleGA.h> // we're going to use the simple GA
#include <ga/GA2DBinStrGenome.h> // and the 2D binary string genome
#include <ga/std_stream.h>
#include "VBF.h"
#define cout STD_COUT
float Objective(GAGenome &); // This is the declaration of our obj function.
int
main(int argc, char **argv)
{
int n = 0, m = 0;
int popsize;
int ngen;
float pmut;
float pcross;
// We generate random seed
for(int ii=1; ii<argc; ii++) {
if(strcmp(argv[ii++],"seed") == 0) {
GARandomSeed((unsigned int)atoi(argv[ii]));
}
}
n = atoi(argv[1]);
m = atoi(argv[2]);
int width = 1 << n;
int height = m;
popsize = atoi(argv[3]);
ngen = atoi(argv[4]);
pmut = atof(argv[5]);
pcross = atof(argv[6]);
GA2DBinaryStringGenome genome(width, height, Objective);
GASimpleGA ga(genome);
ga.populationSize(popsize);
ga.nGenerations(ngen);
ga.pMutation(pmut);
ga.pCrossover(pcross);
ga.evolve();
// Now we print out the best genome that the GA found.
cout << ga.statistics() << "\n";
cout << ga.statistics().bestIndividual() << "\n";
return 0;
}
float
Objective(GAGenome& g) {
using namespace VBFNS;
VBF F;
NTL::mat_GF2 T;
NTL::RR nonlin, lind, suma, nlm, ldm;
NTL::ZZ s, smin, smax;
long spacen, m;
int d, n, ai, maxai;
GA2DBinaryStringGenome & genome = (GA2DBinaryStringGenome &)g;
float score=0.0;
spacen = genome.width();
m = genome.height();
T.SetDims(spacen,m);
for(int i=0; i<spacen; i++){
for(int j=0; j<m; j++){
T[i][j] = to_GF2(genome.gene(i,j));
}
}
F.puttt(T);
nonlin = nl(F);
lind = ld(F);
nlm = nlmax(F);
ldm = ldmax(F);
d = deg(F);
n = logtwo(spacen);
ai = AI(F);
maxai = aimax(F);
s = sigma(F);
smin = sigmamin(F);
smax = sigmamax(F);
suma = nonlin/nlm+lind/ldm+to_RR(d)/to_RR(n)+to_RR(ai)/to_RR(maxai)+1.0-(to_RR(s-smin)/to_RR(smax-smin));
conv(score,suma);
return score;
}
The following program illustrates how can be set the seed (included in a file called “seed.bin” with the binary representation of the Truth Table of the function) of the Genetic algorithm for the o4 objective function:
#include <ga/GASimpleGA.h> // we're going to use the simple GA
#include <ga/GA2DBinStrGenome.h> // and the 2D binary string genome
#include <ga/std_stream.h>
#include "VBF.h"
#define cout STD_COUT
float Objective(GAGenome &); // This is the declaration of our obj function.
int
main(int argc, char **argv)
{
int n = 0, m = 0;
int popsize;
int ngen;
float pmut;
float pcross;
n = atoi(argv[1]);
m = atoi(argv[2]);
int width = 1 << n;
int height = m;
popsize = atoi(argv[3]);
ngen = atoi(argv[4]);
pmut = atof(argv[5]);
pcross = atof(argv[6]);
GA2DBinaryStringGenome genome(width, height, Objective);
ifstream inStream("seed.bin");
if(!inStream){
cerr << "Cannot open " << "seed.bin" << " for input.\n";
exit(1);
}
inStream >> genome;
inStream.close();
GASimpleGA ga(genome);
ga.populationSize(popsize);
ga.nGenerations(ngen);
ga.pMutation(pmut);
ga.pCrossover(pcross);
ga.evolve();
// Now we print out the best genome that the GA found.
cout << ga.statistics() << "\n";
cout << ga.statistics().bestIndividual() << "\n";
return 0;
}
float
Objective(GAGenome& g) {
using namespace VBFNS;
VBF F;
NTL::mat_GF2 T;
NTL::RR nonlin, lind, suma, nlm, ldm;
long spacen, m;
int d, n, ai, maxai;
GA2DBinaryStringGenome & genome = (GA2DBinaryStringGenome &)g;
float score=0.0;
spacen = genome.width();
m = genome.height();
T.SetDims(spacen,m);
for(int i=0; i<spacen; i++){
for(int j=0; j<m; j++){
T[i][j] = to_GF2(genome.gene(i,j));
}
}
F.puttt(T);
nonlin = nl(F);
lind = ld(F);
nlm = nlmax(F);
ldm = ldmax(F);
d = deg(F);
n = logtwo(spacen);
ai = AI(F);
maxai = aimax(F);
suma = nonlin/nlm+lind/ldm+to_RR(d)/to_RR(n)+to_RR(ai)/to_RR(maxai);
conv(score,suma);
return score;
}
11.3. 9-variable Boolean functions with nonlinearity 242¶
Below you can find the Truth Tables of a set of 9-variable functions with the the best known nonlinearity (242). These Boolean functions can be grouped in five different affine equivalence classes:
A representative Boolean function of class f1
A representative Boolean function of class f2
A representative Boolean function of class f3
A representative Boolean function of class f4
A representative Boolean function of class f5
whose Frequency distribution of the absolute values of the Walsh Spectrum are the following:
| f | Values |
|---|---|
| f1 | (4,30),(12,46),(20,226),(28,210) |
| f2 | (4,30),(12,46),(20,226),(28,210) |
| f3 | (4,30),(12,46),(20,226),(28,210) |
| f4 | (4,56),(12,58),(20,154),(28,244) |
| f5 | (4,57),(12,91),(20,97),(28,267) |
whose Frequency distribution of the absolute values of the Autocorrelation Spectrum are the following:
| f | Values |
|---|---|
| f1 | (0,129),(8,298),(16,60),(24,9),(32,2),(40,13),(512,1) |
| f2 | (0,150),(8,196),(16,148),(24,12),(32,5),(512,1) |
| f3 | (0,183),(8,223),(16,84),(24,6),(32,4),(40,10),(56,1),(512,1) |
| f4 | (0,157),(8,232),(16,84),(24,8),(32,17),(40,10),(48,3),(512,1) |
| f5 | (0,192),(8,156),(16,129),(24,9),(32,13),(40,3),(48,6),(64,3),(512,1) |
The following table shows the values some criteria take for the functions within each class:
| Class | LD | DEG | AI | MAXAC |  |
|---|---|---|---|---|---|
| f1 | 118 | 7 | 4 | 40 | 324608 |
| f2 | 120 | 7 | 4 | 32 | 324608 |
| f3 | 114 | 7 | 4 | 56 | 324608 |
| f4 | 116 | 7 | 4 | 48 | 343424 |
| f5 | 112 | 7 | 4 | 64 | 354560 |
11.4. 9-variable Balanced Boolean functions with nonlinearity 240¶
Below you can find the profiles of 9-variable balanced functions with the the best known nonlinearity (240). Each row represents a profile with four values separated by commmas: Algebraic Degree, Algebraic Immunity, Absolute Indicator and Sum-of-square Indicator:
567 different profiles were found with nonlinearity 240 and algebraic degree 8. The algebraic immunity takes values from the set {4, 5}.The linearity distance takes values from the set {110, 112, 114, 116, 118, 120, 122 }. The absolute indicator takes values from the set { 24, 32, 40, 48, 56, 64, 72 }. The sum-of-square indicator takes 137 different values between 323456 and 377600.
11.5. 11-variable Balanced Boolean functions with nonlinearity 992¶
Below you can find the profiles of 11-variable balanced functions with the the best known nonlinearity (992). Each row represents a profile with four values separated by commmas: Algebraic Degree, Algebraic Immunity, Absolute Indicator and Sum-of-square Indicator:
3316 different profiles were found with nonlinearity 992. The algebraic degree takes values from the set { 9, 10 }. The algebraic immunity takes values from the set { 4, 5 }. The absolute indicator takes values from the set: { 120, 128, 136, 144, 160, 168, 176, 192, 200, 208, 224, 232, 240, 248, 256, 264 }. The sum-of-square indicator takes 682 different values between 5244800 and 5844608.